K-Diag: Knowledge-enhanced Disease Diagnosis in Radiographic Imaging
|
|
1CMIC, Shanghai Jiao Tong University
|
2Shanghai AI Laboratory
|
Accepted by MICCAI-BTSD2023 (Workshop Oral)
|
Abstract
In this paper, we consider the problem of disease diagnosis.
Unlike the conventional learning paradigm that treats labels independently, we propose a knowledge-enhanced framework, that enables training visual representation with the guidance of medical domain knowledge.
In particular, we make the following contributions:
First, to explicitly incorporate experts' knowledge, we propose to learn a neural representation for the medical knowledge graph via contrastive learning, implicitly establishing relations between different medical concepts.
Second, while training the visual encoder, we keep the parameters of the knowledge encoder frozen and propose to learn a set of prompt vectors for efficient adaptation.
Third, we adopt a Transformer-based disease-query module for cross-model fusion, which naturally enables explainable diagnosis results via cross attention.
To validate the effectiveness of our proposed framework, we conduct thorough experiments on three x-ray imaging datasets across different anatomy structures, showing our model is able to exploit the implicit relations between diseases/findings, thus is beneficial to the commonly encountered problem in the medical domain, namely, long-tailed and zero-shot recognition, which conventional methods either struggle or completely fail to realize.

Architecture
Overview of the knowledge-enhanced disease diagnosis workflow.
The knowledge encoder (left) is first trained to learn a neural representation of the medical knowledge graph via contrastive learning,
and then used to guide the visual representation learning in our knowledge-enhanced classification model (right).
While training the visual encoder, we keep the parameters of the knowledge encoder frozen and propose to learn a set of prompt vectors for efficient adaptation.
Finally we adopt a Transformer-based disease-query module for cross-model fusion, which naturally enables explainable diagnosis results via cross attention.

Quantitative Results
R1: Analysis of Knowledge-Enhanced Classification Model
Comparison to Conventional Training Scheme.
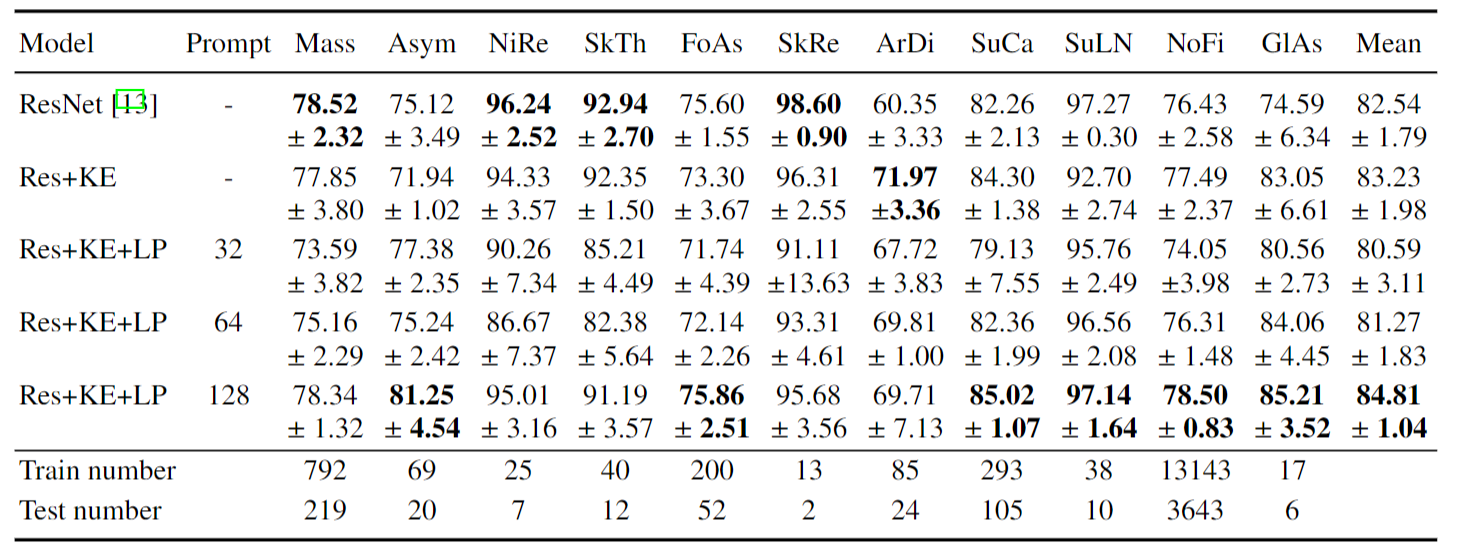
Compare with Baseline Models with ResNet-50 and ViT-16 as backbone on disease classification tasks.
KE indicates the proposed knowledge encoder, LP indicates the proposed learnable prompt module, and the number denotes the prompt number.
AUC scores averaged across different diseases are reported.
Our knowledge-enhanced model achieves a higher average AUC on all datasets across different architectures.
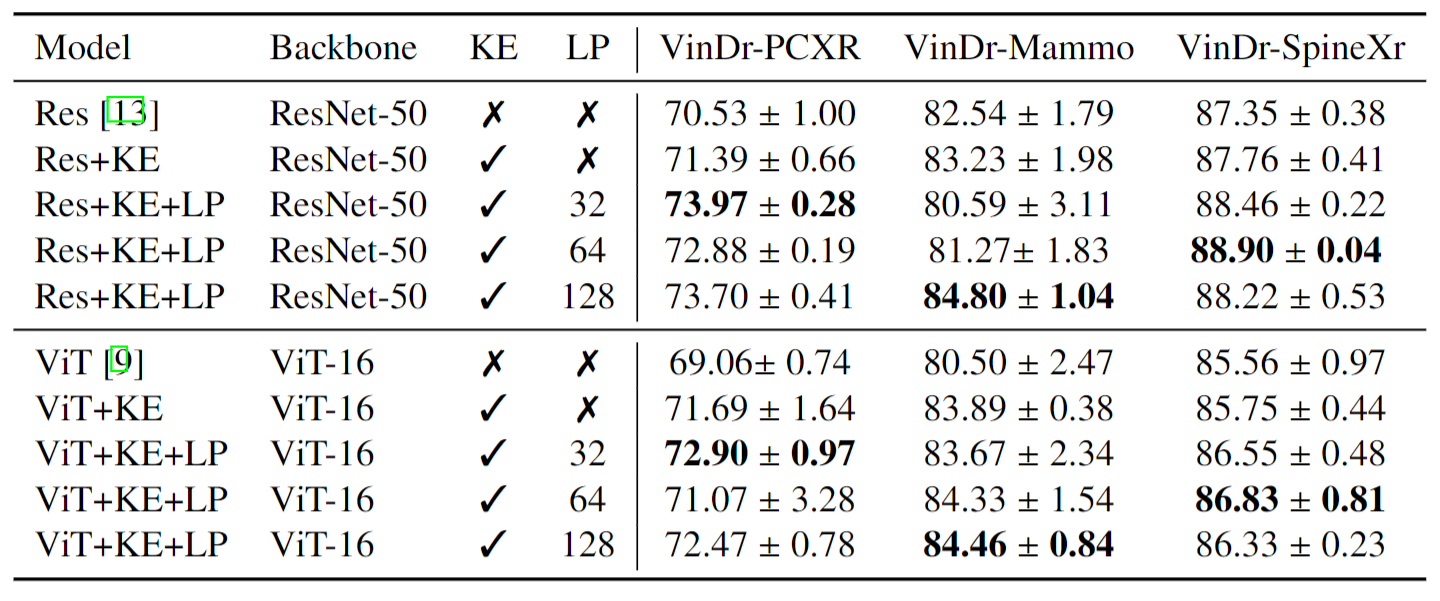
Compare with Baseline Models on VinDr-Mammo Task. AUC scores are reported.
Detail results for each diseases are shown in the table to explain that, on VinDr-Mammo, LP sometimes may have a lower scores mainly because of some classes with extremly few cases.
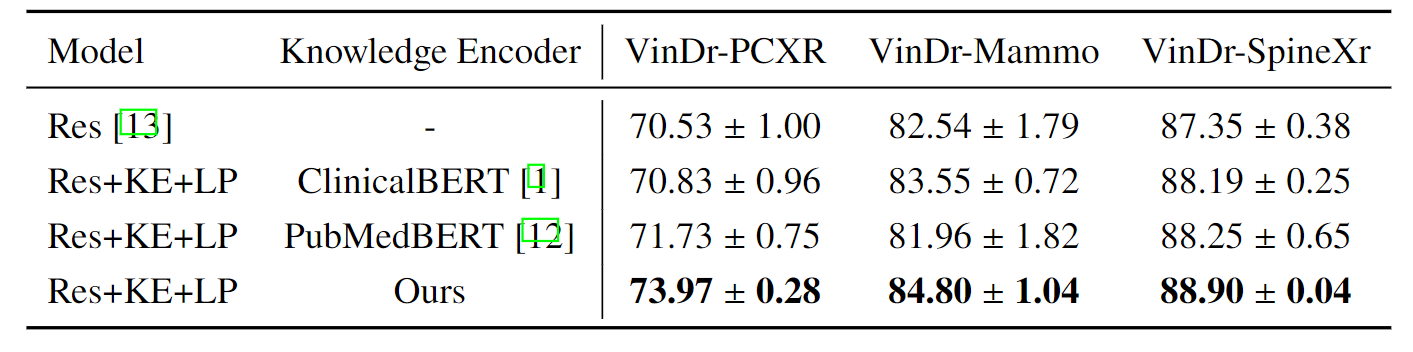
R2: Analysis of the Knowledge-Enhanced Text Encoder
Ablation study on knowledge encoder with ResNet as a backbone, we use the optimal prompt numbers according to the ablation study,
i.e., 32 for VinDr-PCXR, 128 for VinDr-Mammo, and 64 for VinDr-SpineXr.
we can make two observations: (i) guiding visual representation learning with domain knowledge
generally works better, e.g., results of using ClinicalBERT or PubMedBERT outperform
conventional training with discrete labels, (ii) our proposed knowledge-enhanced text
encoder consistently demonstrates superior results, that can be attributed to the explicitly
injected domain knowledge, rather than implicitly learning it from the document corpus.
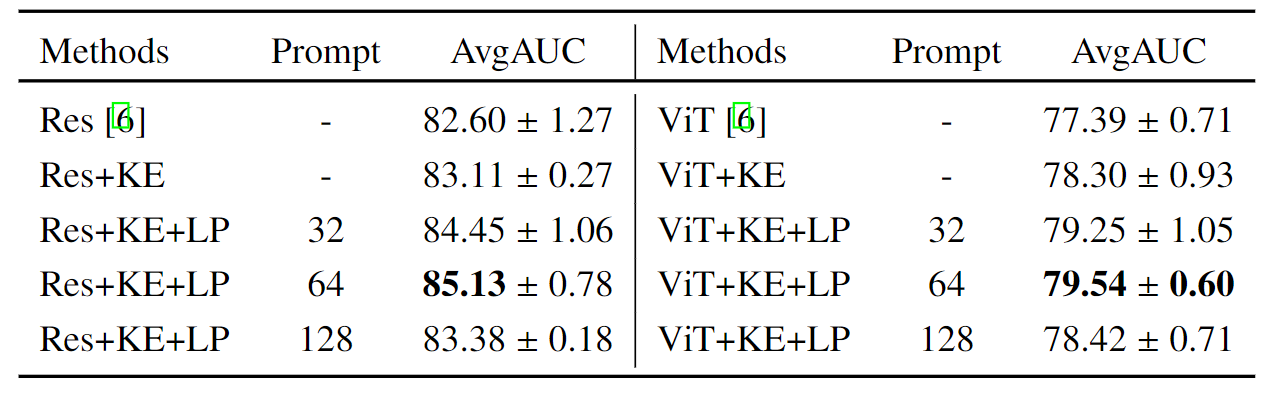
R3: Analysis on the CXR-Mix
The Ability to Combine Various Partial-labeled Datasets:
Compare with Baseline Models on disease classification tasks on the assembling dataset.
AvgAUC refers to the AUC score averaged across different diseases. The first line refers to the use
of the training flow proposed by TorchXrayVision and use ResNet or ViT as the backbone.
Unlike the traditional approach, which requires to carefully merging the label space from different datasets
to benefit from them, our formulation of embedding the 'disease name' with a knowledge
encoder naturally enables us to train models on the mixture of multiple datasets,
handling different granularities of diagnosis targets and inconsistent pathology expression.
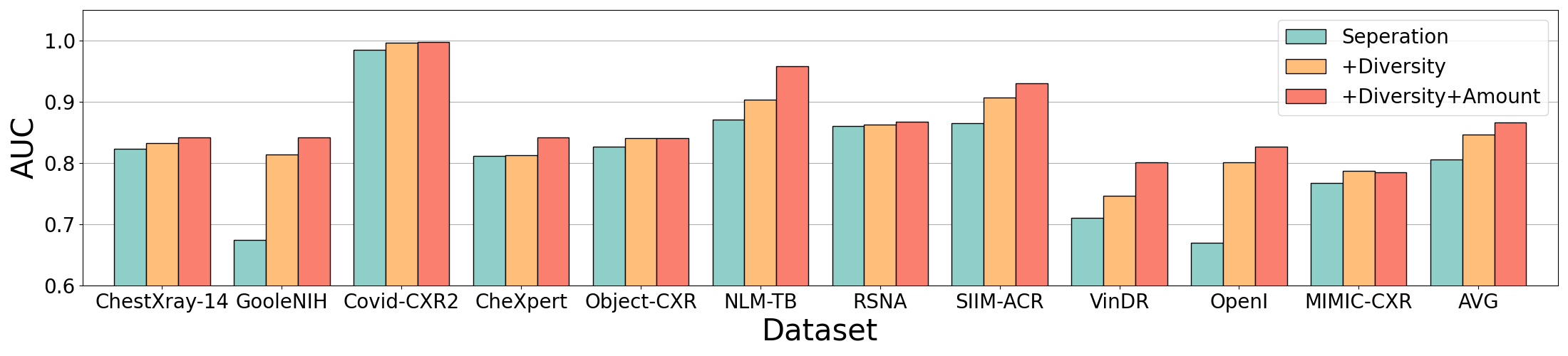
The Ability to Leverage Class Diversity:
Analyse the performance gain on the assembling dataset. "Seperation" refers to using a
single dataset to train our framework. "+Diversity" refers to adding the cases beyond the target
classes, increasing the class diversity, and keeping the data amount of the target classes constant.
"+Diversity+Amount" means directly mixing the 11 datasets and for most datasets, the data
amount of the target classes will further increase.
The Ability to Diagnose Open-set Unseen Diseases:
AUC and 95% CI are shown on the unseen classes under the zero-shot setting. n represents
the number of related cases. Generally, our method achieves an AUC of at least 0.800 on 14
findings, at least 0.700 on 45 findings and at least 0.600 on 79 findings out of 106 radiographic findings where n > 50 in the
PadChest test dataset (n = 39, 053).

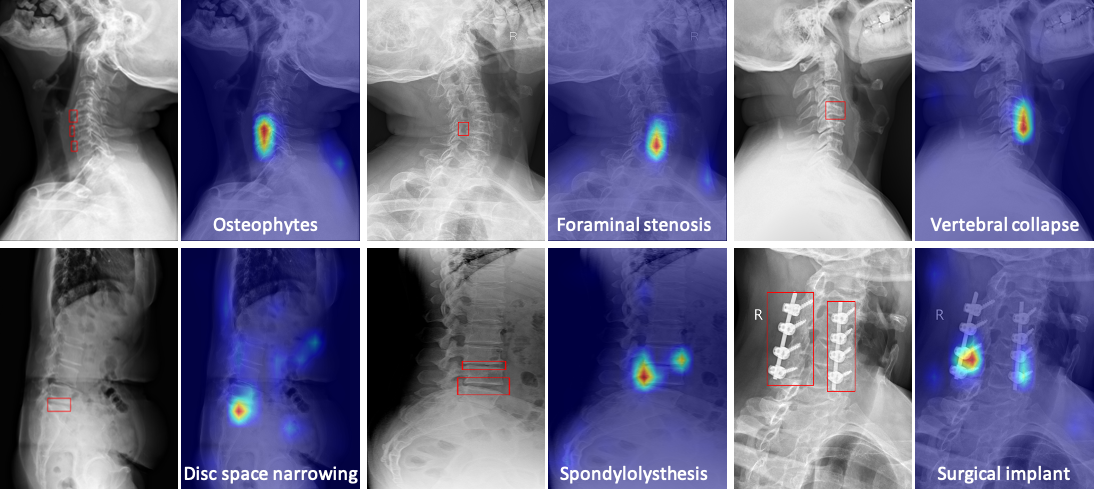
Visualizations of Zero-shot Grounding
We average the cross-attention map in each transformer layer in the disease query module,
and visualize the results in Figure. The model's attention well matches radiologists'
diagnoses of different diseases, i.e. red boxes labeled by radiologists.
Acknowledgements
Based on a template by Phillip Isola and Richard Zhang.